Bulk RNA-seq
Our standard services include genomic alignments, data quality control and normalization, and identification of differentially expressed genes.
For RNA-Seq analysis, we offer:
- FASTQC quality control of raw sequencing reads
- Alignment to standard reference genome using STAR
- Gene-level abundance measurements
- Sample-level unsupervised hierarchical clustering and principle component analysis
- Differential expression testing for pairwise, group comparisons as well as more complex models (multifactorial, time-series, etc) using DESeq2
- Integrative Genome Browser based visualization
| Analysis Type | Features |
|---|---|
| RNA seq | QC: PCA+tSNE+correlation plots demonstrating clustering between biological replicates DE: Volcano Plots, MA Plots, gene expression heat maps showing statistically significant genes showing high fold change |
| Small-RNA seq | QC: PCA+tSNE+correlation plots demonstrating clustering between biological replicates DE: Volcano Plots, MA Plots, gene expression heat maps showing statistically significant genes showing high fold change |
| ChIP-seq | Peak Calling: ChIP-seq peak calling with Macs2 and ENCODE pipelines Coverage Graphs: Creation of genome-wide coverage graphs normalized for sequencing depth and IP efficiency QC: PCA+tSNE+correlation plots demonstrating clustering between biological replicates DE: Volcano Plots, MA Plots, peak region enrichment heat maps showing statistically significant regions showing high fold change |
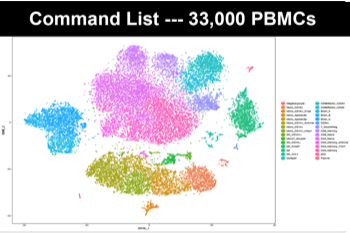
| Single-cell RNA seq | Preprocessing: Support for all data preprocessing steps for the 10x Genomics Single Cell platform, including merging of multiple runs Exploratory Data Analysis: Unsupervised clustering of cells, identification of cluster specific gene markers DE: Identification of differentially expressed genes between conditions (genotypes, treatments) within or between clusters |
| Custom | Stand-alone analysis can be performed based on the researcher's requirements. Contact Walter Eckalbar for more info. |
All analyses are available for download from UCSF's secure Box site. We schedule a meeting at the end of each study to discuss the data analysis in detail.